Summary
1. The ectopic expression of genes has proved to be an extremely valuable tool for biologists. The most widely used systems involve electrically or chemically mediated transfer of genes to immortalized cell lines and, at the other end of the spectrum, transgenic animal models. As would be expected, there are compromises to be made when employing either of these broad approaches. Immortalised cell lines have limited "physiological relevance" and transgenic approaches are costly and out of the reach of many laboratories. There is also significant time required for the de novo generation of a transgenic animal.
2. As a viable alternative to these approaches we describe the use of recombinant adenovirus and Sindbis virus to deliver genes to cells and tissues.
3. We exemplify this approach with studies from our laboratories. i) An investigation of Ca2+
handling deficits in cardiac myocytes of hypertrophied hearts using infection with recombinant adenovirus encoding either green fluorescent protein (GFP) or the sarcoplasmic/endoplasmic reticulum calcium-ATPase (Serca2a). ii) A study of the mechanism of macrophage/microglial migration by infection of embryonic phagocytes with a GFP encoding virus and co-culture with brain slices to then track the movement of labelled cells. ii) We are also exploiting the natural tropism of the Sindbis virus to label neurones in hippocampal brain slices in culture to resolve high-resolution structure and to map neuronal connectivity.
4. Further development of these approaches should open new avenues of investigation for the study of physiology in a range of cells and tissues.
Introduction
Gene transfer, a simple and attractive concept, involves the transfer of DNA to cells of interest. Progress in gene transfer technology has made it a potentially powerful tool for the treatment of a wide variety of diseases (Romano et al., 2000). Early gene transfer techniques using chemical methods such as calcium phosphate or liposomes and physical methods such as electroporation were successfully exploited for basic research but had limited use for gene therapy. Gene transfer technology for gene therapy approaches is based on the ability to efficiently deliver the therapeutic gene to relevant target cells. Efficient delivery in turn is dependent upon type of gene delivery vehicles. Recent advances in gene therapy research and vector technology have led to the development of variety of viral and non-viral vector systems to efficiently deliver genes to cells, tissues and organs by ex vivo and in vivo strategies (Mountain, 2000). Efficient gene transfer systems represent useful tools for basic research and provide new opportunities to study gene function at the cellular and molecular level in a wide variety of cells, tissues, organotypic cultures and whole animals. At this stage integration of gene transfer technology with physiological genomics to study the function of gene products in context of the whole organism and its environment or in a particular cell type at a specific stage of development will play a major role in physiology and medicine.
Gene Transfer Systems
An important challenge to gene transfer technology is the development of a single gene delivery system that can adequately satisfy all of the following criteria: 1) Efficient and targeted cell-specific delivery 2) High levels and long term expression of the transgene 3) Low toxicity for in vivo delivery with minimal side effects 4) Non-immunogenicity. Although existing viral and non-viral vector systems can fulfil some of these criteria, none can single-handedly provide all of the necessary functions. Control of gene expression and targeting to specific cells or tissues is currently an intensive area of gene transfer research. Some of the genetic elements that are being incorporated into the design of new and improved vectors include promoters that are cell-type specific, cell-cycle regulated or tumor selective promoters as well as promoters that respond to radiation, chemotherapy or are heat induced (Nettelbeck et al., 2000).
Non Viral Delivery Systems
Non-viral vectors using mechanical or chemical approaches can efficiently transfect cells in vitro. Mechanical methods involve direct injection or the use of “gene gun technology” to introduce the plasmid DNA (Yang et al., 1996). Low levels of gene expression and inability to use these methods for systemic administration due to the presence of serum nucleases has limited their applications to tissues that are easily accessible such as skin and muscle cells. Electroporation using electrical mediated disruption of cell membranes to effect transfection is used mainly for in vitro applications. Chemical methods are divided into two classes: different formulations of cationic liposomes and cationic polymers such as polylysine, protamine, DEAE dextran or polyethyleneimine (PEI). Classical liposomes (positively charged) have been used to deliver encapsulated drugs and transfer genes into cells in culture (Gao & Huang, 1995). Problems with encapsulation of DNA have led to the development of different formulations of cationic liposomes that are able to interact spontaneously with negatively charged DNA. The transfection efficiency of the liposome/DNA complexes in vivo is very low and can sometimes be cytotoxic in vitro. New and improved formulations of cationic lipids that have enhanced the cellular internalization and transfection efficiency are being used for human gene therapy. The success of non-viral delivery will be greatly dependent on the ability to design systems that can transfect cells with high efficiency, increased stability in presence of serum proteins and reduced toxicity to cells both in vitro and in vivo. One advantage of this system is they have no constraints on size of the gene that can be delivered.
Viral Delivery Systems
Viruses are naturally evolved vehicles that efficiently transfer their genes into host cells. This ability has made them attractive as tools for gene delivery purposes. Viral vectors that have been extensively studied and genetically manipulated for safety concerns in laboratory research and for in vivo gene transfer protocols include retroviruses, adenoviruses, herpes simplex viruses, lentiviruses, adeno associated viruses and Sindbis viruses. Each of the viral vectors has their own individual advantages, problems, and specific applications. Choice of viral vectors is dependent on gene transfer efficiency, capacity to carry foreign genes, tropism, toxicity, stability, immune responses towards viral antigens and potential viral recombination. For functional studies of different transgenes our laboratory is using recombinant adeno and Sindbis viruses for gene transfer purposes and this paper will discuss and review literature related to only these viral gene delivery systems.
Adenoviral Mediated Gene Transfer
Adenoviral genome consists of double stranded linear DNA of ~36 kb in length (Graham & Prevec, 1995). Adenoviral vectors can infect a wide variety of cells and tissues. They can transfer genes to both proliferating and quiescent cells and express the transgene at very high levels. Transgene expression is transient, as these viruses do not integrate into the host genome. To generate replication defective viruses early genes such as E1A, E1B, E2, E3 and E4 involved in adenoviral gene transcription, DNA replication and host cell immune suppression can be deleted. The first-generation recombinant adenoviruses were constructed using a two-plasmid system. The “shuttle” plasmid contains part of the viral genome where the E1A and E1B genes are replaced by a transgene driven by its own regulatory system. The “helper” plasmid contains a complete but unpackageable viral genome. On cotransfection of the two plasmids, homologous recombination takes place to generate a recombinant adenovirus encoding the transgene. Infectious viral particles can be generated in permissive host cells such as HEK293 in which E1A proteins are provided in trans.
First generation recombinant adenoviral vectors have several drawbacks. Transgenes of only 4-5 kb can be packaged. They cause significant cytotoxicity in infected cells and immunological responses in the infected animal. Low efficiency of homologous recombination in HEK293 cells made the process of generating recombinant adenovirus very tedious. To increase the packaging capacity of the foreign gene (~ 7 kb), viral vectors in which E1A and E3 or E4 are deleted were developed (Bett et al., 1995; Gao et al., 1996). To reduce the cytotoxicity in vivo and prolong the transgene expression, a second generation of adenoviral vector with E1 and E2 deletions/mutations was developed (Engelhardt et al., 1994a,b). To simplify the process of generating recombinant viruses, Vogelstein and group (He et al., 1998) developed the AdEasy method that has the significant advantage in that homologous recombination is carried out in E. coli bacterial cells. This made the process of screening for recombinants less cumbersome. Successful recombinants are isolated and then transfected into HEK293 cells to generate replication deficient infectious viral particles. These recent advances in adenoviral technology have made them attractive tools for physiological studies, functional genomics and gene therapy purposes (Wang & Huang, 2000).

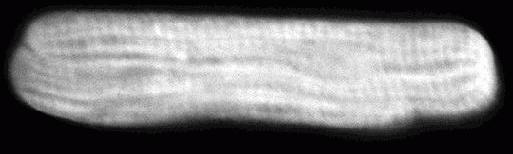
Figure 1. Laser scanning confocal microscopy image of AdSERCA2a-EGFP infected myocyte from rat right ventricle. Expression of EGFP is apparent throughout the cytosol of the cell which exhibits a longitudinal banding pattern reflecting exclusion of expressed protein from the mitochondria, sarcoplasmic reticulum and T-tubules (cell length 120 mm).
Ca2+Handling In Cardiac Hypertrophy
In our laboratory recombinant adenoviruses were generated using the AdEasy system. Within 24 hrs of infection with a recombinant adenovirus carrying GFP, high levels of GFP expression (as evidenced by specific fluorescence emission), was observed in majority (95%) of adult rat cardiac myocytes possessing the rod-shaped morphology characteristic of healthy Ca2+-tolerant cardiac myocytes (Fig. 1). This gene transfer method was then used to manipulate the Ca2+ handling system of isolated cardiac myocytes from rats suffering cardiac hypertrophy and heart failure (Reilly et al., 2001). The rate of Ca2+ sequestration of the sarcoplasmic reticulum (SR) ATP dependent Ca2+ pump is a major determinant of cardiac relaxation and it is clear that reductions in the expression of pumps underlie the prolonged calcium ( Ca2+) transients and consequent reduced contractile performance seen in human cardiac hypertrophy and heart failure. As such, modulation of intracellular Ca2+ levels, Ca2+ kinetics or Ca2+ sensitivity is the focus of many current therapeutic approaches to improve contractile performance in the hypertrophic or failing heart. While there are three highly homologous genes encoding SR Ca2+ pumps, SERCA2a is the specific isoform found in cardiac (and slow-twitch skeletal) muscle, and is abundantly expressed in both atrial and ventricular compartments of mammalian myocardium (Kiriazis & Kranias, 2000).
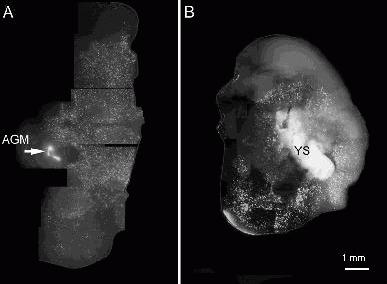
Figure 2. Brain slices from a post-natal day 4 rat pup cultured for 6 days with GFP-labelled embryonic haemopoietic tissues. A) Aorta-gonad-mesonephros, AGM. B) The yolk sac, YS. These tissues where placed in the ventricles of the brain slice. In both cases cells from the haemopoietic tissues have migrated into the brain slice forming a dense distribution.
In freshly isolated cardiac myocytes from rats with monocrotaline-induced right ventricular hypertrophy, Serca2a gene transfer resulted in a marked restoration of Serca2a protein expression levels and completely normalised the timecourse of the stimulated Ca2+ responses in these cells without altering diastolic Ca2+ values or Ca2+ transient amplitudes. These results highlight the importance of Serca2a deficiencies in the hypertrophic phenotype of cardiac muscle and outline a simple, effective viral approach for manipulation and improvement of complex cardiac functions.
Origins of Microglia
We have used adenovirus modified to express GFP to label cells in organotypic co-culture experiments which were designed to establish the developmental potential of embryonic haemopoietic cells in rats. Before the bone marrow develops to become the source of all blood cells a number of transient haemopoietic sites exist at different times in the developing embryo. To determine whether haemopoietic cells of the yolk sac and the aorta-gonad-mesonephros region had the potential to develop into the ramified phagocytic cells in the brain (the microglia) these embryonic tissues were isolated and co-cultured with organotypic brain slices taken from neonatal rats.
Microglia are considered to be the macrophages of the central nervous system and are seen in the developing brain before bone marrow haemopoiesis. Because the neonatal brain slice is known to provide an environment that supports the differentiation of microglia from macrophage-like precursors it was anticipated that the co-culture conditions would allow the potential of the transient embryonic sites to generate microglia cells to be examined. To identify any cells in the brain slice that are derived from the co-cultured yolk sac and AGM, these haemopoietic tissues were infected with GFP-expressing adenovirus after their removal from embryos (but prior to co-culturing with brain slices).
We examined brain slices after 1 week of co-culture and typically found vast numbers of cells derived from the haemopoietic tissues had invaded the brain slice (Fig. 2). Depending on the age of the embryos from which the tissues were taken, both of the embryonic haemopoietic tissues examined had the capacity to populate brain slices with numerous cells of microglial morphology (numerous ramified processes).
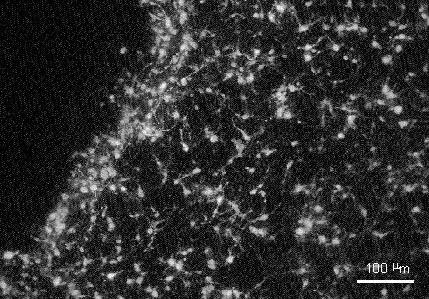
Figure 3. Detail of a region of slice invasion by the GFP labelled cells from the embryonic haemopoietic tissues showing a ramified phenotype typical of microglia.
The adenovirus-mediated GFP labelling allowed detailed examination of the morphology of these cells, enabling us to confirm their ramified state indicative of microglia (Fig. 3). Although we have not confirmed by experiment, our finding of very large numbers of donor tissue derived cells in the slice suggests that considerable proliferation of the GFP labelled population had occurred. If this was the case it would appear that the GFP fluorescence was retained in daughter cells after division.
Adeno-GFP infection of heamopoietic tissues provided intense and apparently robust GFP fluorescence in cells derived from these tissues, allowing the details of the cell morphology to be established. The ease of use and efficacy of this cell labelling method makes it an attractive approach to use in other cell tracking experiments in organ culture or in vivo.
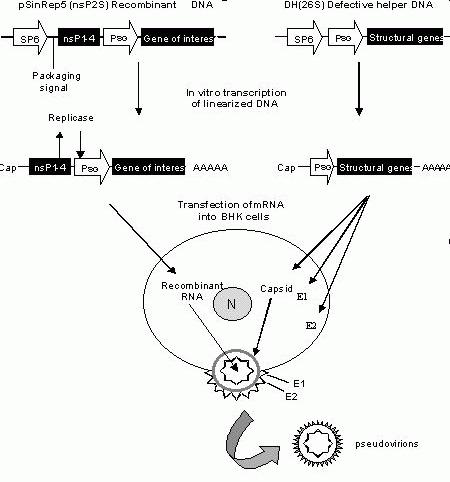
Figure 4. Generation of recombinant Sindbis virus. pSinRep5 (nsps2S) is the recombinant plasmid containing the viral nonstructural genes (nsP1-4) and the subgenomic promoter (PSG) that controls
the transcription of the gene of interest. The defective helper plasmid (DH26S) contains the viral structural genes under the control of the PSG
promoter. In vitro transcription of linearized pSinRep5
and DH26S plasmids results in RNA that has a Cap at the 5’ end and poly A tail at the 3’ end. The transcripts are cotransfected into BHK cells. Translation of the nonstructural genes produces replication enzymes that replicate the recombinant RNA. Capsid protein and E1 and E2 glycoproteins translated from the structural genes of the helper viral RNA, package the recombinant RNA and cause release of the pseudovirions into the medium.
Sindbis Virus mediated Gene Transfer
Sindbis virus is an alphavirus containing a 12-kb single-stranded, positive sense, capped and polyadenylated RNA genome. Introduction of the RNA genome into cells produces infectious virus that is cytopathic. This virus causes encephalitis in mice and rash and arthritis in humans. In mice its primary target is the neurons of the central nervous system (Lustig et al., 1988) and causes neuronal death by inducing apoptosis (Griffin & Hardwick, 1997). Neural infection is dependent on the age of mice and the strain of the virus. The ability to infect a broad range of host cells, small genome size and to amplify and transcribe their genome exclusively in the cytoplasm without affecting host chromosomal machinery are some of the properties that have made Sindbis viruses attractive tools for reengineering purposes and gene transfer. Genetic manipulation of the viral genome, use of two vector system and development of packaging cell lines stably transformed with inducible structural protein expression cassette (Polo et al., 1999) has resulted in new Sindbis vectors that are noncytopathic in mammalian cells, express the transgene at very high levels and reduced the generation of contaminating replication competent virus.
We generated recombinant Sindbis virus using the two-vector system (Fig 4). Briefly, the plasmid pSinRep5 (nsP2S) is used to generate recombinant RNA molecules for transfection and infection. This plasmid contains the viral nonstructural protein genes (nsP1-4) required for replicating RNA transcripts in vivo (in the cell), the SP6 promoter for in vitro transcription, packaging signal and subgenomic promoter (PSG) for transcription of the subgenomic RNA containing the gene of interest. To reduce the cytopathic effects of the Sindbis virus a single mutation (P726S) has been introduced in the nsP2 gene (Dryga et al., 1997). The defective helper plasmid DH(26S) that provides the structural proteins in trans contains the SP6 promoter for in vitro transcription and the PSG promoter for transcription of the viral capsid and E1 and E2 genes. Both the pSinRep5 (nsp2S) and DH(26S) plasmids are linearized and then in vitro transcribed to generate corresponding mRNA that has a Cap at the 5’end and a poly A tail at the 3’end. The transcripts are cotransfected into BHK cells. In the cell cytoplasm, the recombinant RNA is translated to produce the replication enzymes that synthesize the recombinant RNA. Translation of the structural genes from the defective helper RNA produce the capsid protein and E1 and E2 glycoproteins that package the recombinant RNA and subsequently cause release of the viral particles into the medium.
Neuronal structure revealed by sindbis-GFP.
Recombinant Sindbis virus carrying the GFP gene efficiently infects neuronal cells from hippocampus of neonatal rat brain (Fig. 5). Brain slices were grown on cell culture inserts (Millipore) for 1-2 days and then microinjected with the Sindbis virus. About 14-18 hours following infection GFP fluorescence was visualised in the cell body and the dendritic processes of the neurons. In many cases nerve processes could be traced for several millimetres suggesting that this technique can be used for tracing long-range nerve projections in whole brain. Further support for this approach comes from a recent report (Chen et al., 2000), that employed in vivo injection of Sindbis-EGFP to obtain high-resolution images of neurones both in slices and in vivo. These studies thus suggest the potential use of this virus for studying dynamic changes and neural connectivity during development.
Viral Transfer For Physiological Studies - The Future
A number of other exciting possibilities are currently under exploration in our own laboratories including the use of in vivo viral infection to deliver genes to organs and tissues in whole animals. Using this approach our goal is to create genetically modified animals to study the physiological consequences of gene expression. The use of antisense genes may delete endogenous gene expression, akin to a "knockout" animal. Dominant mutant genes that cause disease in humans can be expressed in animals and the phenotype assessed, similar to the approach employed by transgenic and knockin models. Such studies are not only less expensive and more rapid to develop but may also be used as a screening method for the creation of useful genetically modified animal models. Furthermore the infection with multiple dominant mutant genes may be used to examine the more difficult polygenic disorders such as hypertension and mental disease.
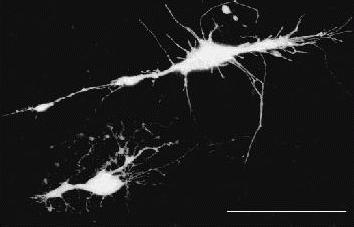
Figure 5. Hippocampal neurones infected with Sindbis encoding GFP. Tissue slices were made from a post natal day 7 rat brain and cultured overnight. Recombinant Sindbis (50 nl of pseudovirions) was injected into the slice using a Drummond nanoject II and the slices examined on a Zeiss confocal LSM510 after 24 hrs in culture. For this image 20 confocal planes were projected onto the Z-axis. Scale bar is 50 μm.
A final application under study in our laboratories is the use of viral vectors for the in vivo delivery of genetically encoded reporter genes. We have employed two such sensors in our own studies, cameleons (Miyawaki et al., 1997, 1999) and camgaroos (Baird et al., 1999). Both these are GFP-based sensors that convert Ca2+ levels into readily measurable fluorescence light signals. We have modified them for targeting to organelles or cellular compartments of interest (Petrou et al., 2000) that obviates the need for high spatial resolution and results in a commensurate gain in temporal resolution.
The completion of the first draft of the human genome sequence heralded a new era in genomics and for physiology, opened new vistas. While the sequence and basic function of many genes will soon be resolved, elucidation of the physiological role of these genes is a major new challenge facing modern physiologists. The science of Physiological Genomics is ultimately concerned with solving this challenge and we envisage that viral mediated gene transfer provides physiologist with an important tool for this emerging discipline.
References
Baird, G.S., Zacharias, D.A. & Tsien, R.Y. (1999) Circular permutation and receptor insertion within green fluorescent proteins. Proceedings of the National Academy of Sciences USA, 96, 11241-11246.
Bett, A.J., Krougliak, V. & Graham, F.L. (1995) DNA sequence of the deletion/insertion in early region 3 of Ad5 dl309. Virus Research, 39, 75-82.
Chen, B.E., Lendvai, B., Nimchinsky, E.A., Burbach, B., Fox, K. & Svoboda, K. (2000) Imaging high-resolution structure of GFP-expressing neurons in neocortex In vivo. Learning and Memory, 7, 433-441.
Dryga, S.A., Dryga, O.A. & Schlesinger, S. (1997) Identification of mutations in a Sindbis virus variant able to establish persistent infection in BHK cells: the importance of a mutation in the nsP2 gene. Virology, 228, 74-83.
Engelhardt, J.F., Litzky, L. & Wilson, J.M. (1994a) Prolonged transgene expression in cotton rat lung with recombinant adenoviruses defective in E2a. Human Gene Therapy, 5, 1217-1229.
Engelhardt, J.F., Ye, X., Doranz, B. & Wilson, J.M. (1994b) Ablation of E2A in recombinant adenoviruses improves transgene persistence and decreases inflammatory response in mouse liver. Proceedings of the National Academy of Sciences USA, 91, 6196-6200.
Gao, G.P., Yang, Y. & Wilson, J.M. (1996) Biology of adenovirus vectors with E1 and E4 deletions for liver-directed gene therapy. Journal of Virology, 70, 8934-8943.
Gao, X. & Huang, L. (1995) Cationic liposome-mediated gene transfer. Gene Therapy, 2, 710-722.
Graham, F.L. & Prevec, L. (1995) Methods for construction of adenovirus vectors. Molecular Biotechnology, 3, 207-220.
Griffin, D.E. & Hardwick, J.M. (1997) Regulators of apoptosis on the road to persistent alphavirus infection. Annual Review of Microbiology, 51, 565-592.
He, T.C., Zhou, S., da Costa, L.T., Yu, J., Kinzler, K.W. & Vogelstein, B. (1998) A simplified system for generating recombinant adenoviruses. Proceedings of the National Academy of Sciences USA, 95, 2509-2514.
Kiriazis, H. & Kranias, E.G. (2000) Genetically engineered models with alterations in cardiac membrane calcium-handling proteins. Annual. Review of Physiology, 62, 321-351.
Lustig, S., Jackson, A.C., Hahn, C.S., Griffin, D.E., Strauss, E.G. & Strauss, J.H. (1988) Molecular basis of Sindbis virus neurovirulence in mice. Journal of Virology, 62, 2329-2336.
Miyawaki, A., Griesbeck, O., Heim, R. & Tsien, R.Y. (1999) Dynamic and quantitative Ca2+ measurements using improved cameleons. Proceedings of the National Academy of Sciences USA, 96, 2135-2140.
Miyawaki, A., Llopis, J., Heim, R., McCaffery, J.M., Adams, J.A., Ikura, M. & Tsien, R.Y. (1997) Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature, 388, 882-887.
Mountain, A. (2000) Gene therapy: the first decade. Trends in Biotechnology, 18, 119-128.
Nettelbeck, D.M., Jerome, V. & Muller, R. (2000) Gene therapy: designer promoters for tumour targeting. Trends in Genetics, 16, 174-181.
Petrou, S., Bowser, D.N., Nicholls, R.A., Panchal, R.G., Smart, M.L., Reilly, A.M. & Williams, D.A. (2000) Genetically targeted calcium sensors enhance the study of organelle function in living cells. Clinical and Experimental Pharmacology and Physiology, 27, 738-744.
Polo, J.M., Belli, B.A., Driver, D.A., Frolov, I., Sherrill, S., Hariharan, M.J., Townsend, K., Perri, S., Mento, S.J., Jolly, D.J. et al. (1999) Stable alphavirus packaging cell lines for Sindbis virus and Semliki Forest virus-derived vectors. Proceedings of the National Academy of Sciences USA, 96, 4598-4603.
Reilly, A.M., Petrou, S., Panchal, R.G. & Williams, D.A. (2001) Restoration of calcium handling properties of adult cardiac myocytes from hypertrophied hearts (submitted).
Romano, G., Michell, P., Pacilio, C. & Giordano, A. (2000) Latest developments in gene transfer technology: achievements, perspectives, and controversies over therapeutic applications. Stem Cells, 18, 19-39.
Wang, I. & Huang, I. (2000) Adenovirus technology for gene manipulation and functional studies. Drug
Discovery Today, 5, 10-16.
Yang, N.S., Sun, W.H. & McCabe, D. (1996) Developing particle-mediated gene-transfer technology for research into gene therapy of cancer. Molecular Medicine Today, 2, 476-481.